Eccentricity Centrality
Definition
The greatest distance between v and any other vertex.
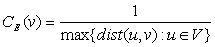 [HAGE, P. 1995]
[HAGE, P. 1995]
The eccentricity is a node centrality index. The eccentricity of a node v is calculated by computing the shortest path between the node v and all other nodes in the graph, then the longest shortest path is chosen (let (v,K) where K is the most distance node from v). Once this path with length dist(v,K) is identified, its reciprocal is calculated (1/dist(v,K)). By doing that, an eccentricity with higher value assumes a positive meaning in term of node proximity. Indeed, if the eccentricity of the node v is high, this means that all other nodes are in proximity. In contrast, if the eccentricity is low, this means that there is at least one node (and all its neighbors) that is far form node v. Of course, this does not exclude that several other nodes are much closer to node v. Thus, eccentricity is a more meaningful parameter if is high. Notably, high and low values are more significant when compared to the average eccentricity of the graph G calculated by averaging the eccentricity values of all nodes in the graph.
Note:The other names are "Harary Graph Centrality" and "Jordan Centrality"
The eccentricity of a node in a biological network, for instance a protein signaling network, can be interpreted as the easiness of a protein to be functionally reached by all other proteins in the network. Thus, a protein with high eccentricity, compared to the average eccentricity of the network, will be more easily influenced by the activity of other proteins (the protein is subject to a more stringent or complex regulation) or, conversely could easily influence several other proteins. In contrast, a low eccentricity, compared to the average eccentricity of the network, could indicate a marginal functional role (although this should be also evaluated with other parameters and contextualized to the network annotations). [SCARDONI, G.]
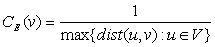
The eccentricity is a node centrality index. The eccentricity of a node v is calculated by computing the shortest path between the node v and all other nodes in the graph, then the longest shortest path is chosen (let (v,K) where K is the most distance node from v). Once this path with length dist(v,K) is identified, its reciprocal is calculated (1/dist(v,K)). By doing that, an eccentricity with higher value assumes a positive meaning in term of node proximity. Indeed, if the eccentricity of the node v is high, this means that all other nodes are in proximity. In contrast, if the eccentricity is low, this means that there is at least one node (and all its neighbors) that is far form node v. Of course, this does not exclude that several other nodes are much closer to node v. Thus, eccentricity is a more meaningful parameter if is high. Notably, high and low values are more significant when compared to the average eccentricity of the graph G calculated by averaging the eccentricity values of all nodes in the graph.
Note:The other names are "Harary Graph Centrality" and "Jordan Centrality"
The eccentricity of a node in a biological network, for instance a protein signaling network, can be interpreted as the easiness of a protein to be functionally reached by all other proteins in the network. Thus, a protein with high eccentricity, compared to the average eccentricity of the network, will be more easily influenced by the activity of other proteins (the protein is subject to a more stringent or complex regulation) or, conversely could easily influence several other proteins. In contrast, a low eccentricity, compared to the average eccentricity of the network, could indicate a marginal functional role (although this should be also evaluated with other parameters and contextualized to the network annotations). [SCARDONI, G.]
Requirements
Require connected and strongly connected network.
Software
- CentiBiN
http://centibin.ipk-gatersleben.de/ - CentiLib
http://centilib.ipk-gatersleben.de/ - CentiScaPe
http://www.cbmc.it/~scardonig/centiscape/centiscape.php - Functional Genomics Assistant (FUGA)
http://code.google.com/p/fuga - GraphStream
http://graphstream-project.org/ - igraph
http://igraph.org - Interference
http://www.cbmc.it/~scardonig/interference/Interference.php - JUNG
http://jung.sourceforge.net - neo4j
http://neo4j.com/ - NetworkAnalyzer
http://med.bioinf.mpi-inf.mpg.de/networkanalyzer/ - SBEToolbox
https://github.com/biocoder/SBEToolbox/releases - sna
http://CRAN.R-project.org/package=sna - SocNetV
http://socnetv.sourceforge.net/ - Visone
http://visone.info/ - Wolfram
http://www.wolfram.com
References
- CARTER T. BUTTS (2014). sna: Tools for Social Network Analysis. R package version 2.3-2. http://CRAN.R-project.org/package=sna
- HAGE, P. & HARARY, F. 1995. Eccentricity and centrality in networks. Social Networks, 17, 57-63.
DOI: 10.1016/0378-8733(94)00248-9


- CSARDI, G. & NEPUSZ, T. 2006. The igraph software package for complex network research. InterJournal, Complex Systems, 1695. [http://igraph.org]
- JUNG, the Java Universal Network/Graph Framework [http://jung.sourceforge.net]
- SCARDONI, G., LAUDANNA, C., TOSADORI, G., FABBRI, F. & FAIZAAN, M. CentiScaPe: Network centralities for Cytoscape. http://www.cbmc.it/~scardonig/centiscape/CentiScaPefiles/CentralitiesTutorial.pdf
- SCARDONI, G., PETTERLINI, M. & LAUDANNA, C. 2009. Analyzing biological network parameters with CentiScaPe. Bioinformatics, 25, 2857-2859.
DOI: 10.1093/bioinformatics/btp517

